Customized Assays Made Easy with scanR TruAI
With the ongoing development of sample preparation protocols, fluorescent markers, and the various cell lines used in screening, it is difficult to provide robust, ready-to-go analysis protocols for a broad range of analysis applications (e.g., apoptosis, autophagy, cell count, cell differentiation, mitotic index, protein expression, and transmigration). As a result, in labs where many different applications or non-standard assays are expected, having the tools to build your own assay is a major advantage.
To achieve this, the Olympus scanR high-content screening (HCS) system uses an assay builder inspired by flow cytometry. Detected objects are displayed in parameter scatter plots where clouds of populations emerge, revealing phenotypes that can be displayed in galleries for visual inspection and gated for statistical quantification (Figures 1 and 2). The analysis parameters can be easily tuned to cover various applications, including kinetic assays.
This same approach based on scatter plots, galleries, and gates is ideal to judge the robustness of deep learning models for image segmentation, a field that is revolutionizing microscopy.
In this webinar, you’ll get an introduction to the scanR HCS system and learn how to:
- Build your own assays with a live demonstration of the workflow
- Use the same tools to validate and judge the performance of neural networks for image analysis
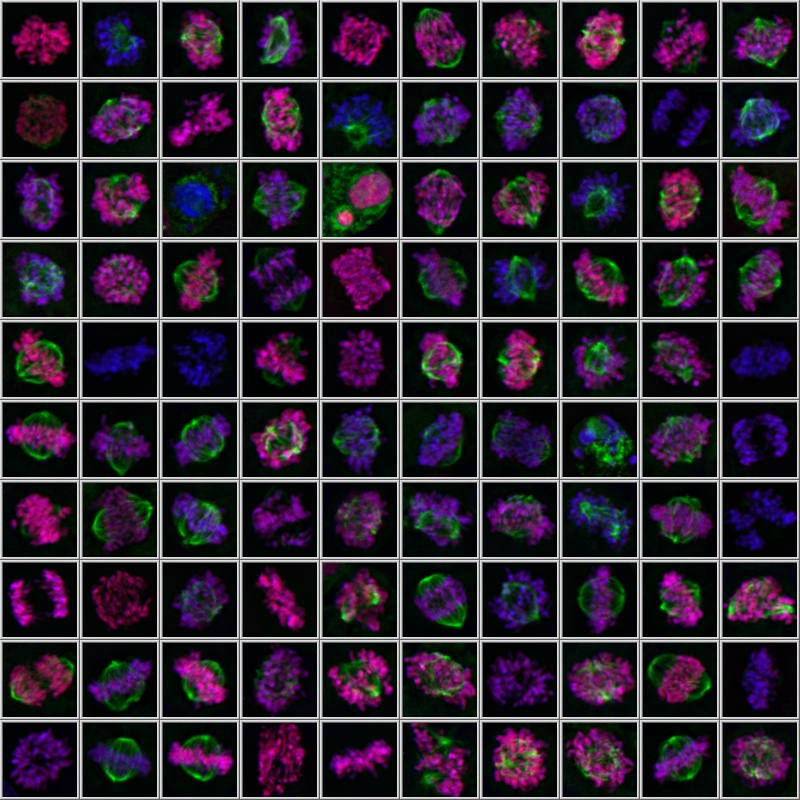 Figure 1. Gallery of a gate of mitotic cells measured with a spinning disk | 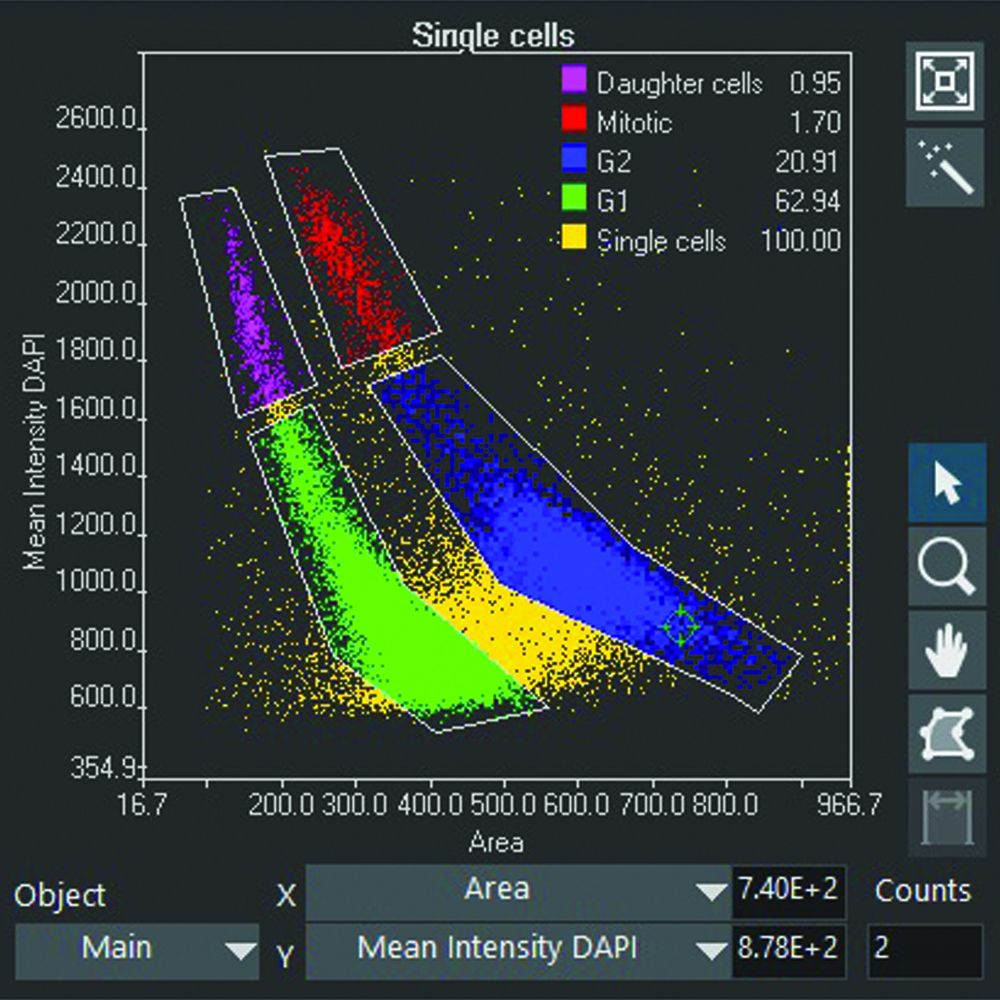 Figure 2. Gated cell populations | 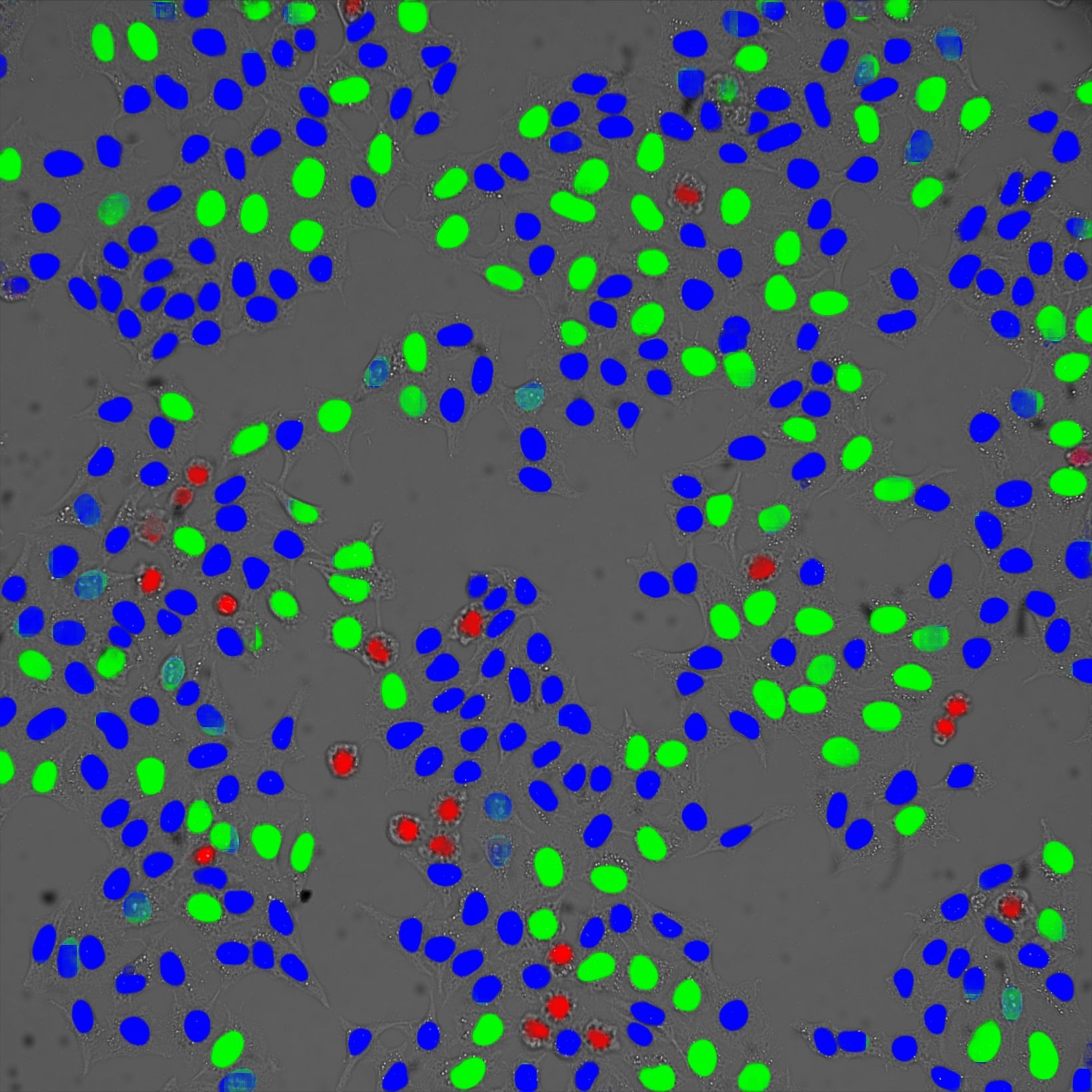 Figure 3. Cell cycle classification in brightfield based on deep learning |
Presenter:
 | Manoel Veiga |
Register now:
|
对不起,此内容在您的国家不适用。