TruAI技术是一种由人工智能驱动的强大的图像分析工具,适用于生命科学和材料科学软件。利用深度神经网络,它可以开发出专为特定成像应用定制的人工智能模型。Evident于2019年通过scanR高内涵筛选站推出了这一技术,后来又将其推广到更多产品中。
TruAI技术可在PC机上本地运行,用户无需具备编码或编程技能。开发的AI模型是可以在Evident的分析软件包之间轻松交换的文件,包括scanR Analysis(scanR分析)、cellSens Count and Measure( cellSens计数和测量)和VS200-Detect(VS200-探测)。
TruAI技术在以下三个关键应用领域展露出优势:
- 图像分析中的目标分割和分类
- 图像采集工作流程中的样品检测
- 图像处理,包括去噪和增强处理
在这篇博文中,我们回顾了TruAI技术在这些生命科学软件包中的演变,展现了其在过去六年中持续改进的历程。
1. 灵活的工具箱(2019年)
TruAI技术的初始版本:2019年的scanR和2020年的cellSens,侧重于为研究人员提供一个灵活的工具箱,用于创建由人工智能驱动的语义分割模型。
训练人工智能模型时,用户首先要在图像中标注真实情况,然后再将标注的图像加载到训练界面中。该模型对标注的图像进行分析,并在迭代过程中根据注释优化预测结果。
早期TruAI工具箱的主要功能包括:
- 手动注释:用于在图像中自由标注真实情况的标注工具。
- 自动注释: 将现有的分割情况转换为注释。
- 部分标注: 将训练限制在标注的区域,无需对整个图像进行标注。
- 灵活的通道和Z层组合: 可使用单通道或Z层执行简单任务,同时允许自由组合来完成复杂应用。
- 训练进度监控:训练界面自动将数据分成训练数据集和验证数据集,并可实时查看人工智能预测结果。
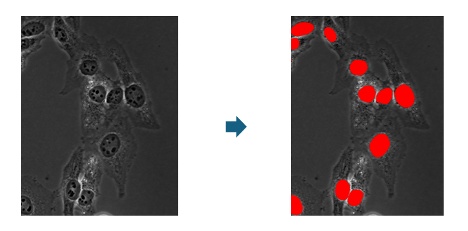
2. 语义分割(2019年)
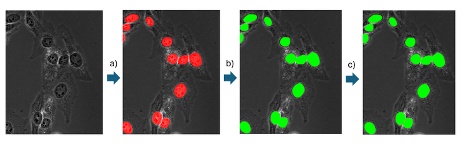
人工智能模型经过训练后,可被应用于各种Evident分析软件包中的新图像(推理)。在语义分割中,人工智能模型会创建一种像素概率图,标出很有可能属于前景的像素。在进行最终目标检测时,会为概率图设置一个阈值,然后采用分水岭分割等经典分割算法进行分割。
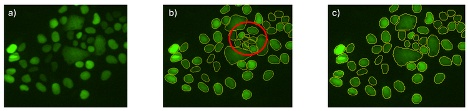
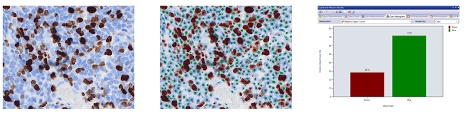
3. 分类(2021年)
单一人工智能模型可以分割图像中的所有目标,并根据这些目标的测量参数(如面积和荧光强度)进行分类。
另外,还可以将多个人工智能模型应用于同一张图像,在不提取测量参数的情况下进行分类,例如,一个人工智能模型只检测处于状态1的细胞,另一个人工智能模型只检测处于状态2的细胞。
这两种功能在TruAI技术的初始版本中就已经成为可能。
然而,分类模型特指单一人工智能模型从背景中区分多个前景类别的能力。而最终目标检测是通过对单一人工智能模型呈现的两个或多个概率应用阈值来实现的。
为了保持灵活性,用户可以在训练过程中决定类别是否可以重叠。
使用分类模型的主要优点是,在探测到许多类别或没有明确的可测量分类特征时,可以简化分析。
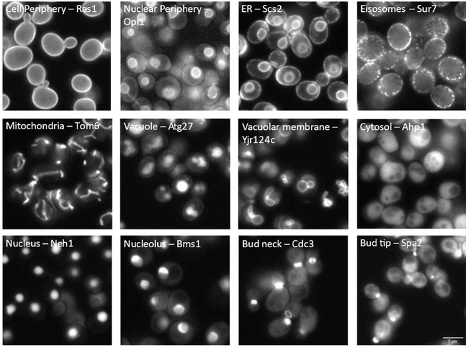
图2:酵母中荧光标记蛋白的细胞区室定位。单一TruAI模型可以根据蛋白质定位对细胞进行分类。要了解更多信息,请参阅我们的应用说明:使用TruAI深度学习技术对酵母蛋白定位进行分类。
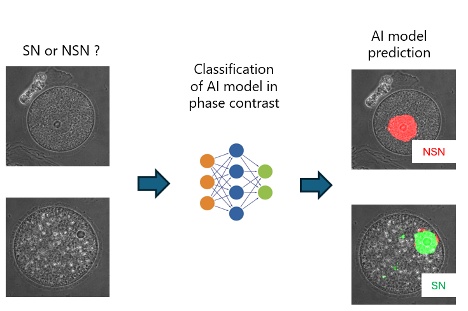
图3:TruAI技术可创建分类人工智能模型用于难以通过人眼进行分类的情况,例如可从未染色的样品中选择高质量的卵母细胞用于体外受精。点击此处,阅读这篇文章,了解更多信息。
4. 实例分割(2021年)
与语义分割人工智能模型不同,实例分割模型可以一步直接分割最终目标,省去了为概率图设置阈值和额外分割的需要。
这种方法简化了工作流程(比较图4和图1),尤其适用于经典分割算法难以分离探测到的目标的情况,如细胞密度较高时。
5. 扩展(2021年、2023年)
人工智能模型的视场有限,通常只有几百个像素,因此对像素分辨率非常敏感。扩展技术可以减轻这些限制。
在训练过程中使用扩展功能(2021年)
当目标超出人工智能模型的视场范围时,模型可能无法识别完整的目标边界,导致检测效率低下。为了解决这个问题,训练中可以加入一个扩展因子,以有效地扩大模型的视场。这种训练方法尤其适用于检测整个大型物体,如斑马鱼、类器官或组织样品中的较大区域。
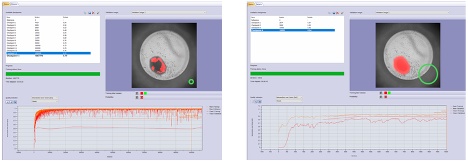
图6. 训练如何检测孔板中处于转化状态的大型类器官。AI模型的视场由绿圈表示。左图:未使用扩展功能的训练,即使经过数千次迭代,所得到的结果也很差。右图:使用25%扩展功能的训练,仅需几百次迭代就能获得高质量的结果。
推理过程中使用扩展功能(2023年)
如果使用10X物镜训练一个人工智能模型,却将其应用于使用40X物镜拍摄的图像,那么检测效率就会很低。TruAI技术的扩展功能,可使用户根据人工智能模型的分辨率调整图像分辨率,以获得优化性能。此外,当实例分割模型导致过度分割时,降低分辨率通常可以改善对整个目标的检测。
6. 实时人工智能(2021年)
实时人工智能是cellSens软件的一个功能,可实时应用人工智能模型,而无需采集图像。人工智能概率或最终目标分割直接显示在实时图像上,可快速进行质量控制检查,在量化应用中进行快速计数,以及在需要对样品进行选择(如选出优质的细胞、精子或卵母细胞)的流程中轻松进行目标识别。
Related Videos
图8. 实时人工智能的推理工作流程。可将具有分类功能的实例分割模型应用于孔板。当用户浏览孔板时,实时图像的左下方会显示概率图和三种细胞类别的计数。
7. 预训练模型(2021年–2024年)
对于没有时间训练自己的人工智能模型的用户,Evident推出了预训练模型,开箱即可使用。以下分析软件包提供了不同类型的预训练人工智能模型:
- scanR分析: 用于明场中的细胞核、细胞、斑点和结构的模型
- 计数和测量以及VS200-Detect:用于细胞核、细胞、IHC细胞分类(Ki-67分析)和微弱全样品识别的模型
- cellSens FV(FV4000: 用于去噪的模型
此外,预训练模型可用作注释的起点。预训练模型的预测结果可被转换为注释,而且用户可以对其进行修正。该工作流程与交互式培训结合使用的效果特别好(参见第9部分)。
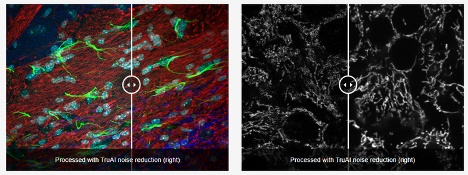
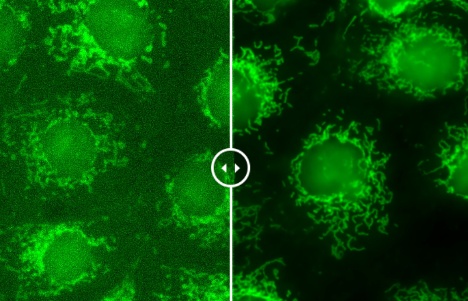
图10. 使用FLUOVIEW FV4000共聚焦显微镜捕获的共振扫描图像(每张图像的左侧)和通过TruAI降噪功能得到增强的图像(每张图像的右侧)。共振扫描仪成像技术能以较低的损伤有效捕捉细胞的高速动态变化。然而,这通常是以较低的信噪比为代价的。TruAI降噪技术通过使用基于SilVIR探测器噪声模式的预训练人工智能模型,可在不影响时间分辨率的情况下增强这些图像。这些经过预训练的TruAI降噪算法既可用于实时处理,也可用于后处理。
8. 自动样品检测(2022年)
人工智能模型可被集成到采集工作流程中,进行自动样品检测,从而节省了整个组织扫描、特定区域识别或罕见事件识别的时间。工作流程通常从低倍率的概览扫描开始,然后再使用更多通道、更多Z层,甚至采用从宽视野到共聚焦转盘显微镜的模式转换,以更高倍率进行详细扫描。
Related Videos
图11. 视频展示了cellSens软件中用于宽视场显微镜的宏观到微观成像的工作流程。
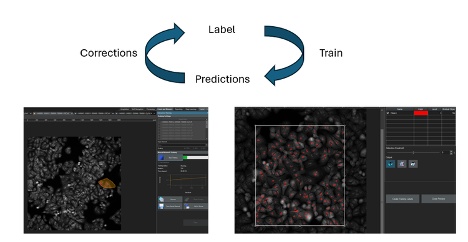
9. 交互式训练(2023年)
2023年推出了一种用于训练人工智能模型的新型交互式工作流程。这种工作流程可使用户以交互方式为图像添加注释、训练模型、查看人工智能预测结果、进行修正,并将修正结果作为新标签用于进一步训练。然后,用户可以继续注释新图像,反复不断地完善模型。通过这种方法,可以快速开发出用于简单应用的人工智能模型。
10. 图像增强(2023年)
用户既可以为分割任务又可以为图像处理操作,训练人工智能模型。例如,可以教授模型使用去噪或反卷积等处理技术。
由于训练是跨通道进行的,因此生成用于训练的真实数据非常简单。例如,为去噪模型创建真实数据非常简单,只需收集两个通道的数据即可:一个具有高信噪比(作为真实数据),另一个使用短曝光和弱光激发。
这种方法的一个主要优势是,研究人员可以使用自己的样品训练人工智能模型,有助于确保其特定目标结构的优化性能,同时还可大幅减少幻觉等伪影现象(虚假、无法解释的结构)。
TruAI技术的持续演进
在过去的六年里,TruAI技术不断发展,在分割、分类、扩展、实时分析和图像增强方面都取得了进步。其灵活性、易用性以及与Evident软件平台的集成,使其成为生命科学领域人工智能驱动图像分析的强大工具。
要详细了解TruAI模型在实践中的工作原理, 请联系我们的显微镜专业团队,申请个性化演示。reach out to our team of microscopy professionals for a personalized demo.